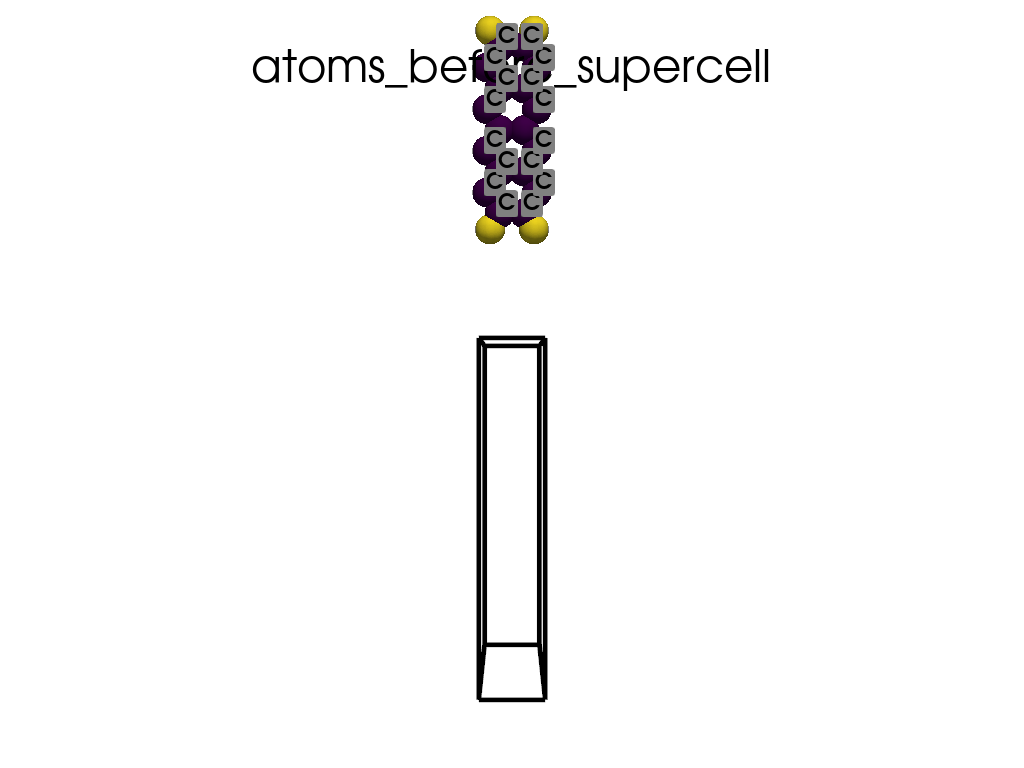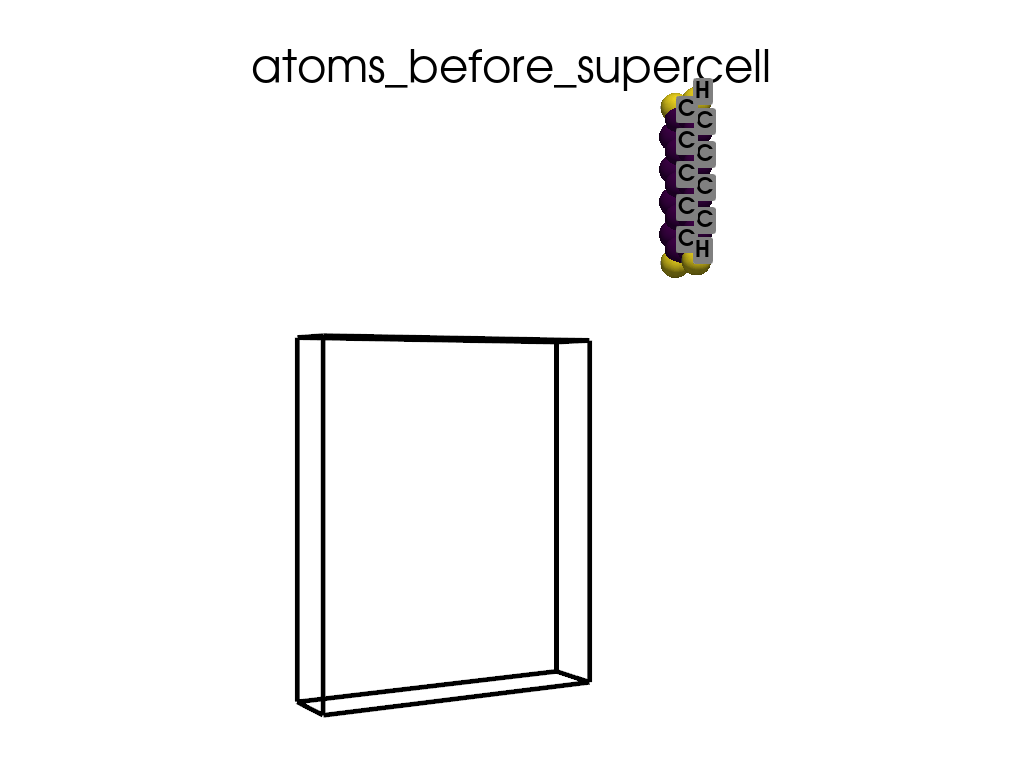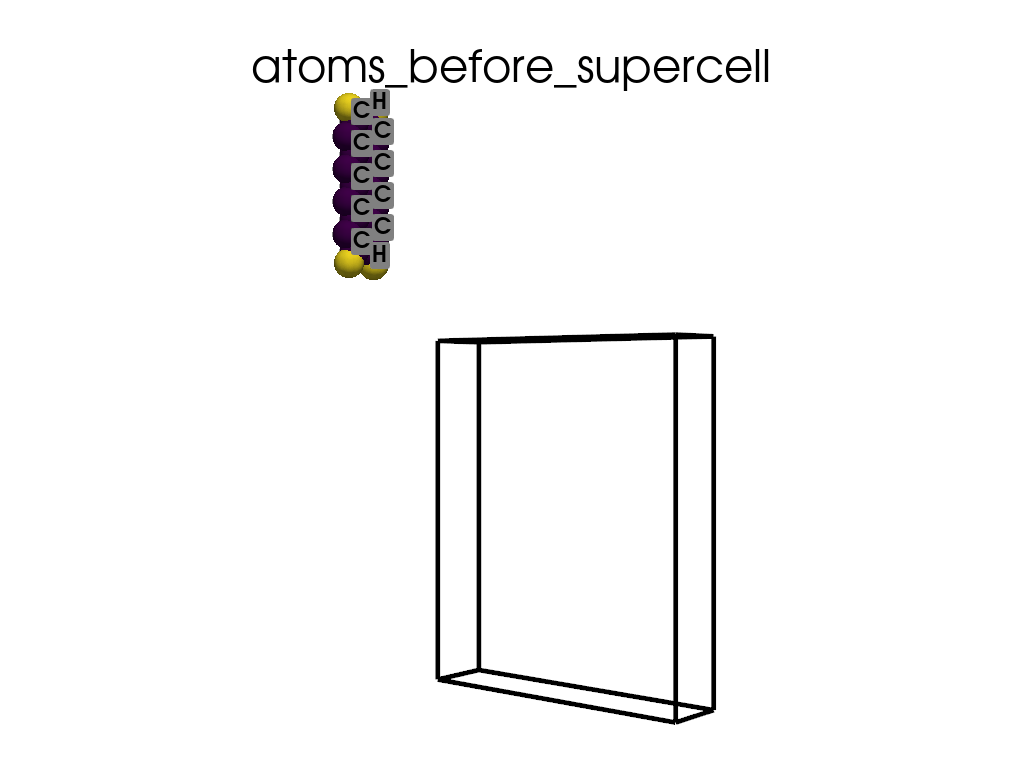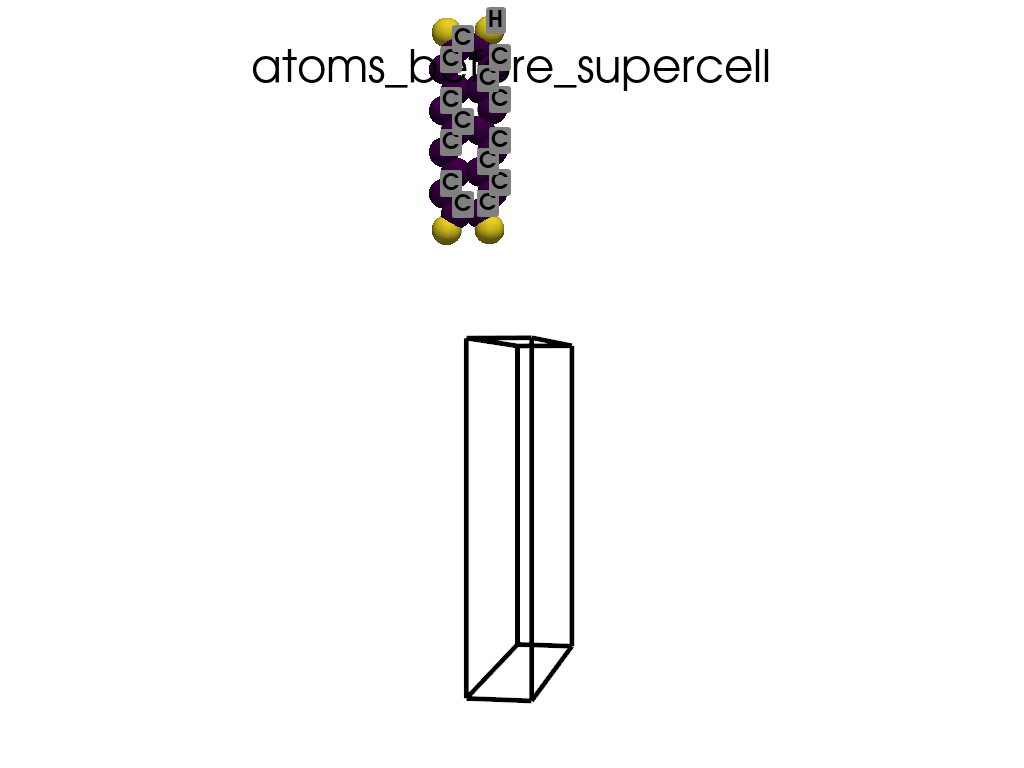Note
Go to the end to download the full example code.
Modifying a POSCAR File: Scaling, Supercells, and Defects#
In this example, we’ll demonstrate several modifications on a POSCAR file using the pyprocar package:
Scaling the lattice vectors to reduce vacuum space.
Generating a supercell.
Introducing defects by changing atom types.
Let’s get started!
import os
from itertools import product
import numpy as np
import pyvista as pv
import pyprocar.pyposcar as p
from pyprocar.utils import DATA_DIR
data_dir = os.path.join(DATA_DIR, "examples", "PyPoscar", "01-poscar_utils")
# You do not need this. This is to ensure an image is rendered off screen when generating exmaple gallery.
pv.OFF_SCREEN = True
Utility function for creating GIF visualizations#
def create_gif(atoms, labels, unit_cell, save_file):
plotter = pv.Plotter()
title = save_file.split(os.sep)[-1].split(".")[0]
plotter.add_title(title)
plotter.add_mesh(
unit_cell.delaunay_3d().extract_feature_edges(),
color="black",
line_width=5,
render_lines_as_tubes=True,
)
plotter.add_point_labels(
points=atoms.points, labels=labels, show_points=False, always_visible=True
)
plotter.add_mesh(
atoms,
scalars="atoms",
point_size=30,
render_points_as_spheres=True,
show_scalar_bar=False,
)
path = plotter.generate_orbital_path(n_points=36)
plotter.open_gif(os.path.join(data_dir, save_file))
plotter.orbit_on_path(path, write_frames=True, viewup=[0, 0, 1], step=0.05)
plotter.close()
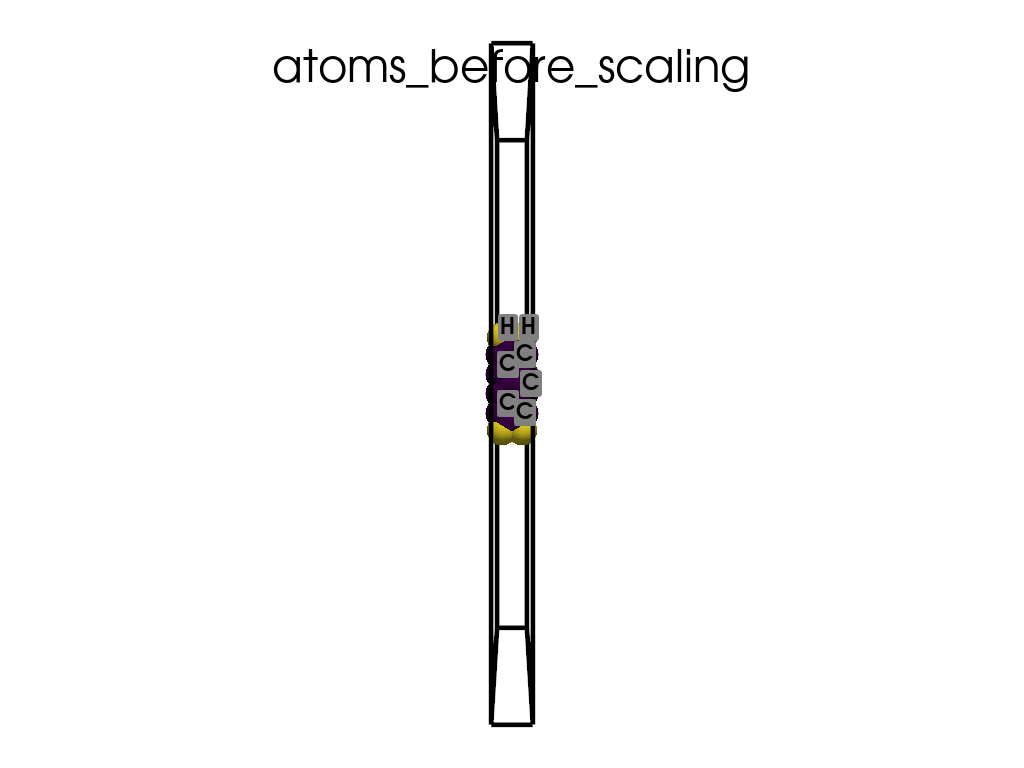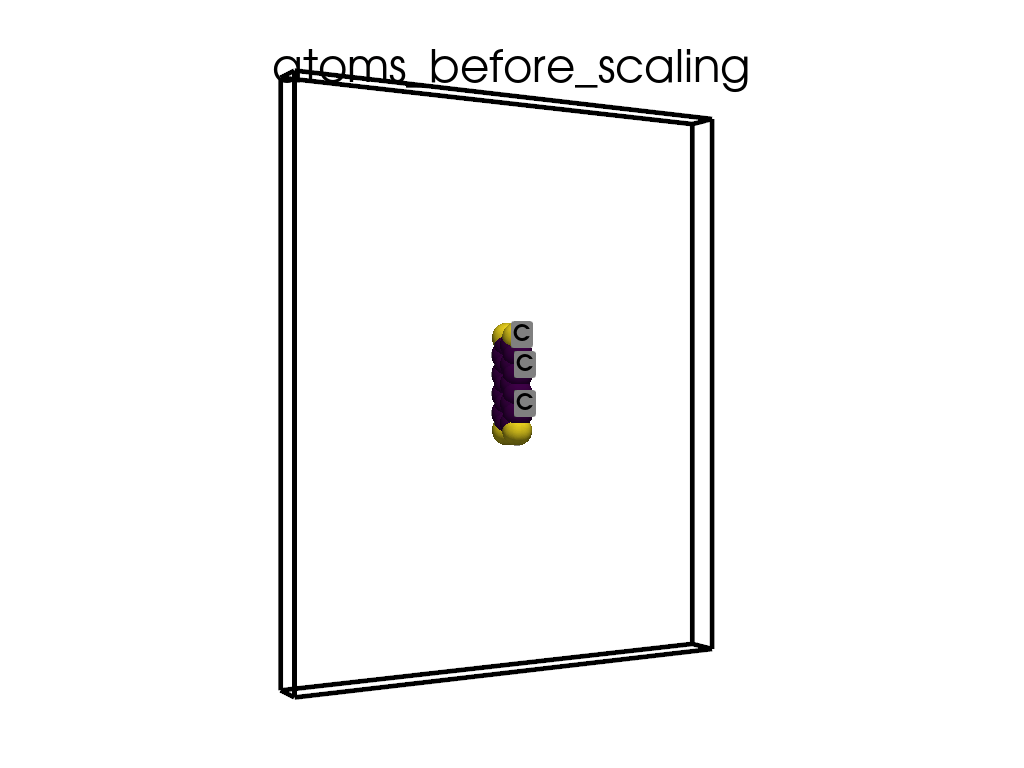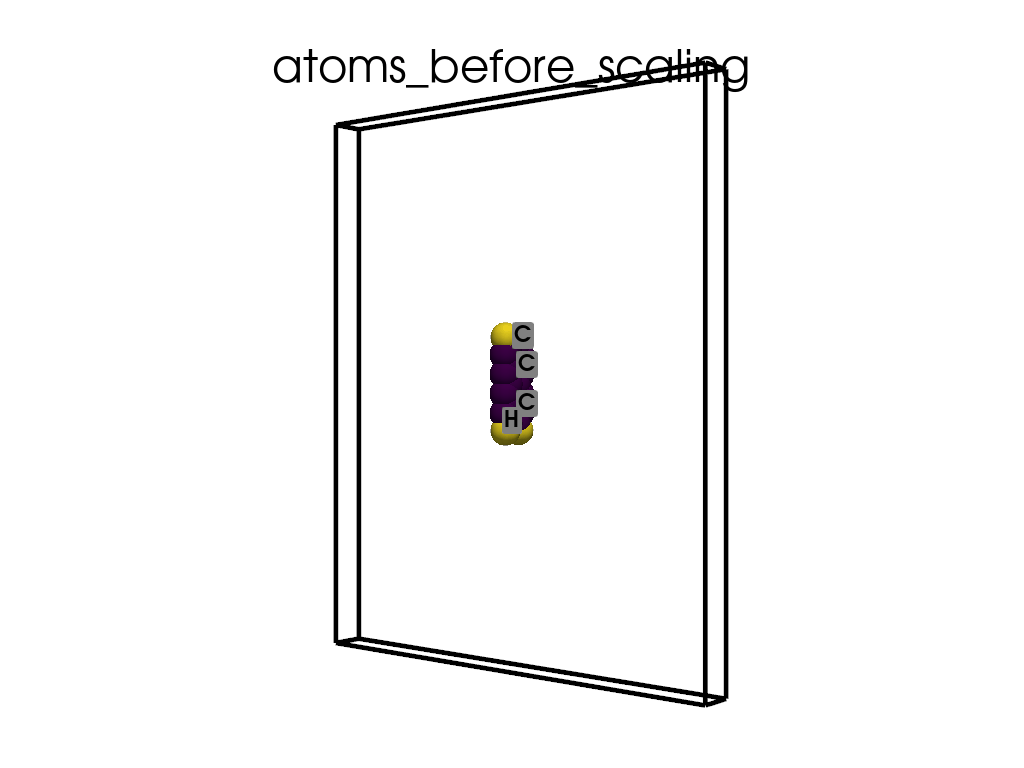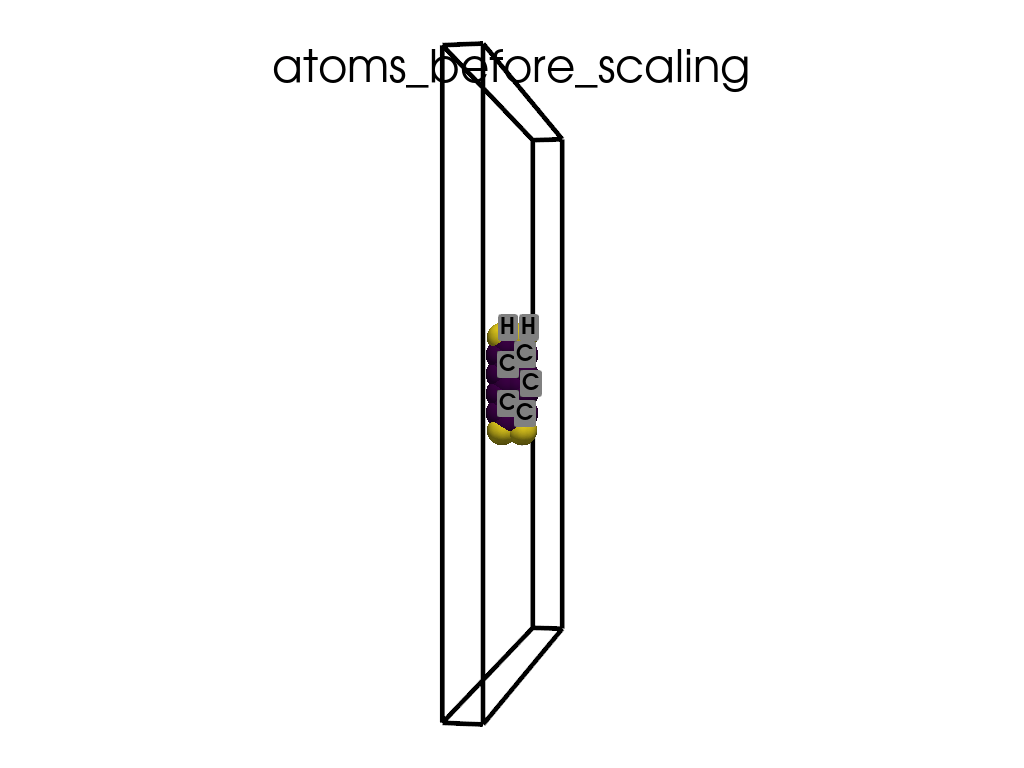
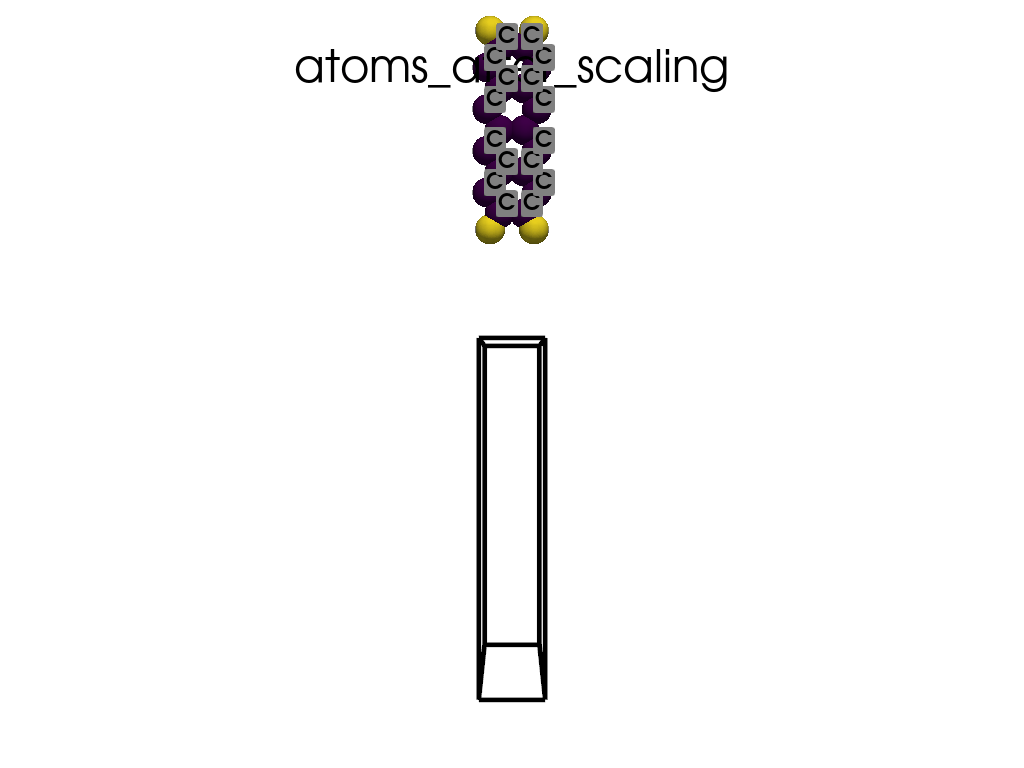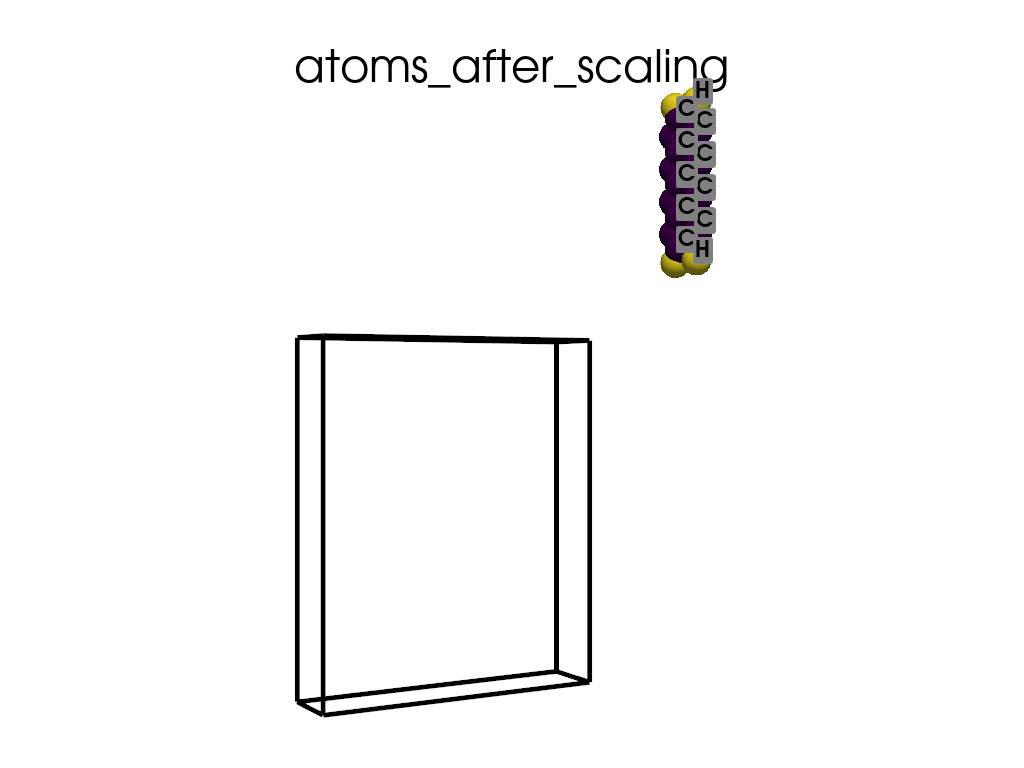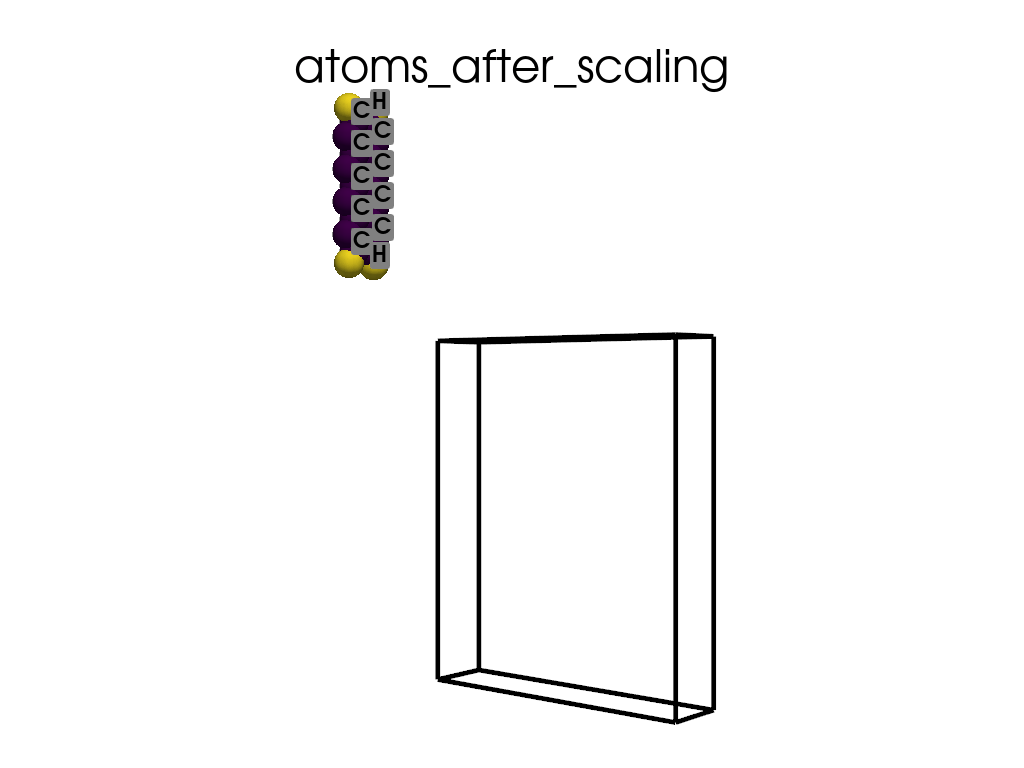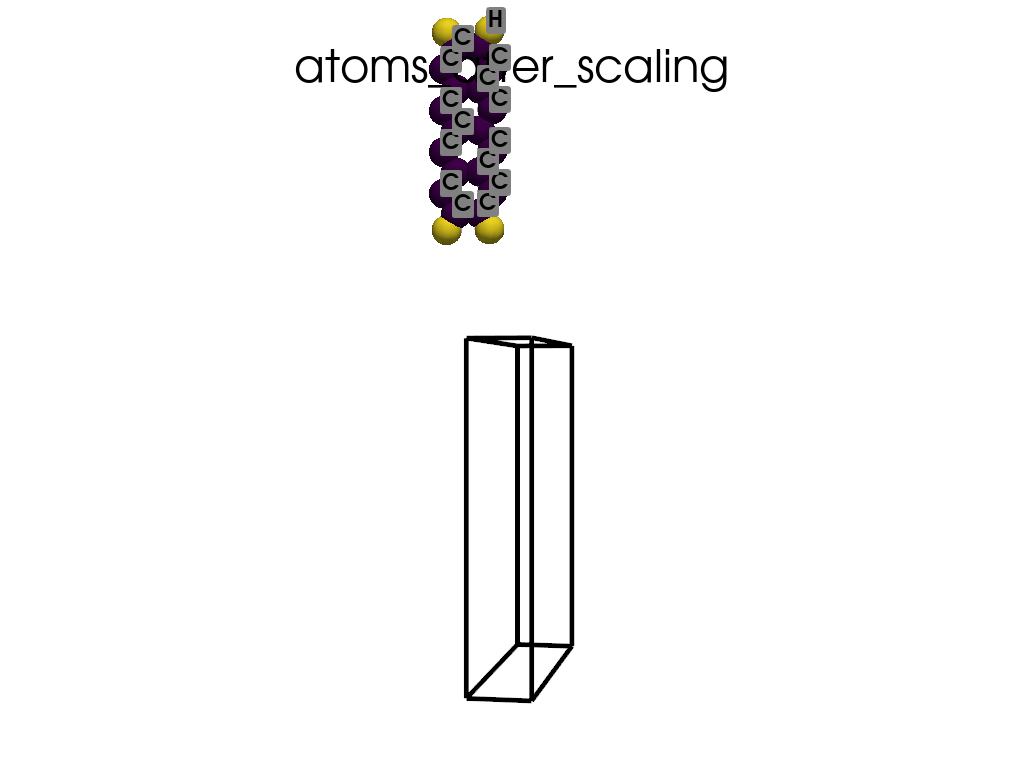
Scaling Vacuum Space in the Lattice#
a = p.poscarUtils.poscar_modify(
os.path.join(data_dir, "POSCAR-9AGNR.vasp"), verbose=False
)
print("the lattice has too much vacuum space\n", a.p.lat)
print("I will shrink these vector by 1/3")
scaling = np.array([1, 1 / 3, 1 / 3])
a.scale_lattice(factor=scaling, keep_cartesian=True)
a.write(os.path.join(data_dir, "POSCAR-9AGNR-smaller.vasp"))
print("New lattice\n", a.p.lat)
tmp_a = p.Poscar(os.path.join(data_dir, "POSCAR-9AGNR.vasp"))
tmp_a.parse()
# Convert positions to Cartesian coordinates for visualization
atoms_before = pv.PolyData(np.dot(tmp_a.dpos, tmp_a.lat))
atoms_before["atoms"] = tmp_a.elm
labels_before = [elm for elm, point in zip(tmp_a.elm, tmp_a.dpos)]
# Define the unit cell using lattice vectors
unit_cell_comb = list(product([0, 1], repeat=3))
unit_cell = np.array(
[
comb[0] * tmp_a.lat[0] + comb[1] * tmp_a.lat[1] + comb[2] * tmp_a.lat[2]
for comb in unit_cell_comb
]
)
unit_cell_before = pv.PolyData(unit_cell)
tmp_a = p.Poscar(os.path.join(data_dir, "POSCAR-9AGNR-smaller.vasp"))
tmp_a.parse()
# Convert positions to Cartesian coordinates for visualization
atoms_after = pv.PolyData(np.dot(tmp_a.dpos, tmp_a.lat))
atoms_after["atoms"] = tmp_a.elm
labels_after = [elm for elm, point in zip(tmp_a.elm, tmp_a.dpos)]
# Define the unit cell using lattice vectors
unit_cell_comb = list(product([0, 1], repeat=3))
unit_cell = np.array(
[
comb[0] * tmp_a.lat[0] + comb[1] * tmp_a.lat[1] + comb[2] * tmp_a.lat[2]
for comb in unit_cell_comb
]
)
unit_cell_after = pv.PolyData(unit_cell)
create_gif(
atoms=atoms_before,
labels=labels_before,
unit_cell=unit_cell_before,
save_file="atoms_before_scaling.gif",
)
create_gif(
atoms=atoms_after,
labels=labels_after,
unit_cell=unit_cell_after,
save_file="atoms_after_scaling.gif",
)
the lattice has too much vacuum space
[[ 4.3878 0. 0. ]
[ 0. 60. 0. ]
[ 0. 0. 71.726 ]]
I will shrink these vector by 1/3
New lattice
[[ 4.3878 0. 0. ]
[ 0. 20. 0. ]
[ 0. 0. 23.9087]]
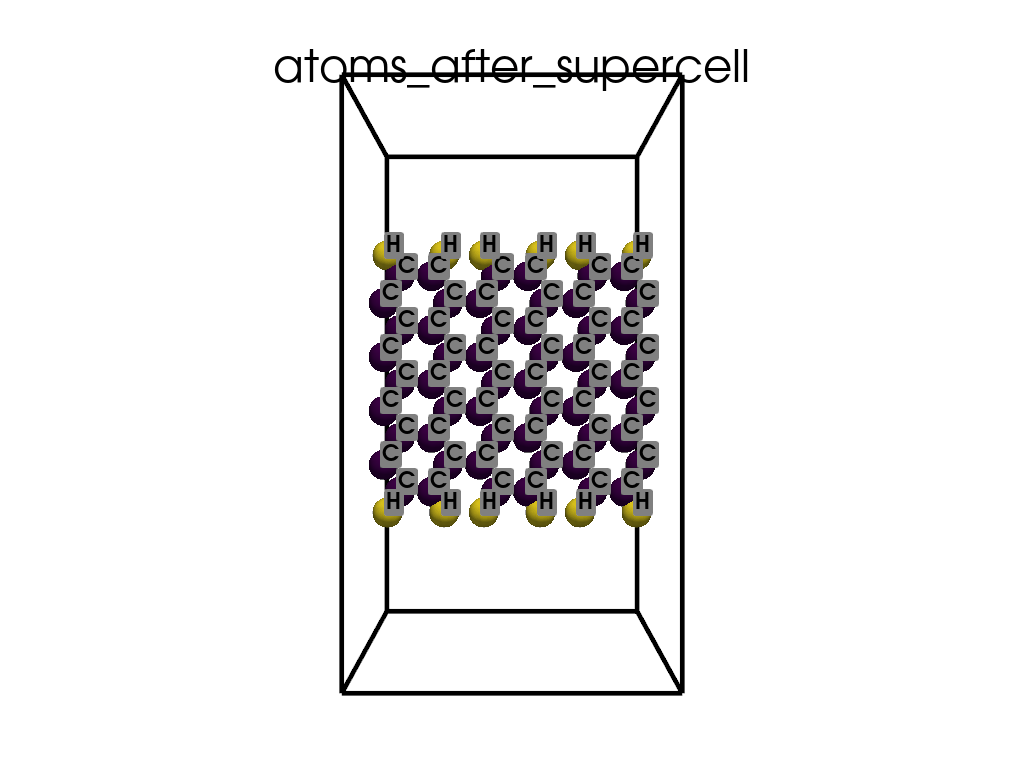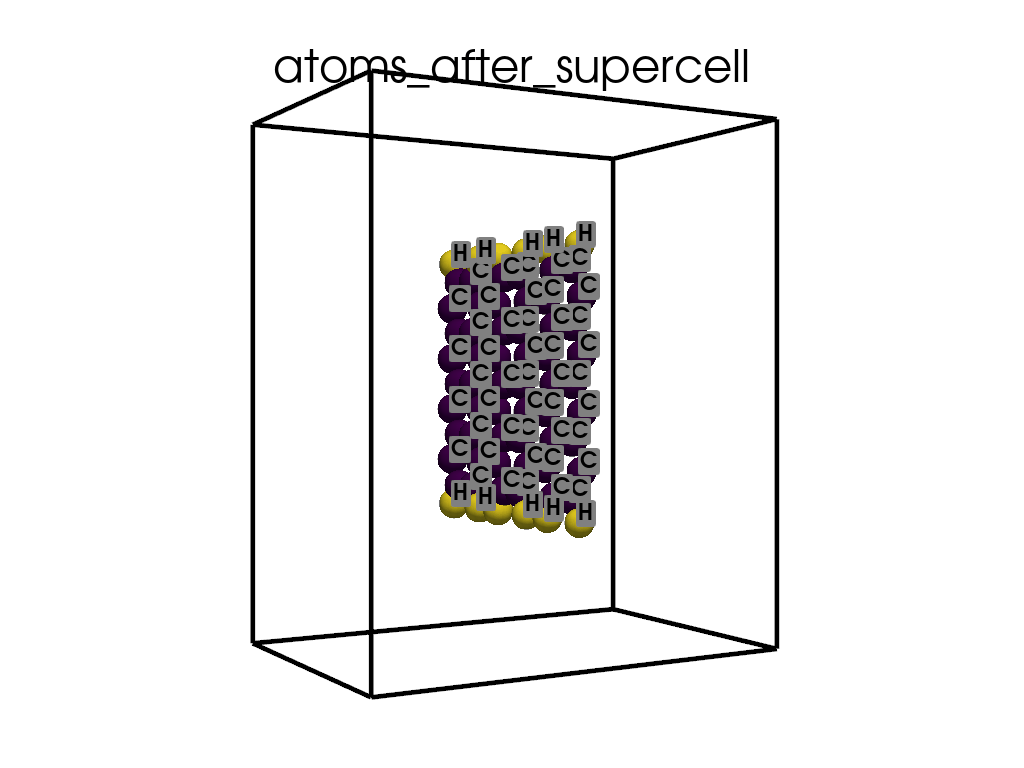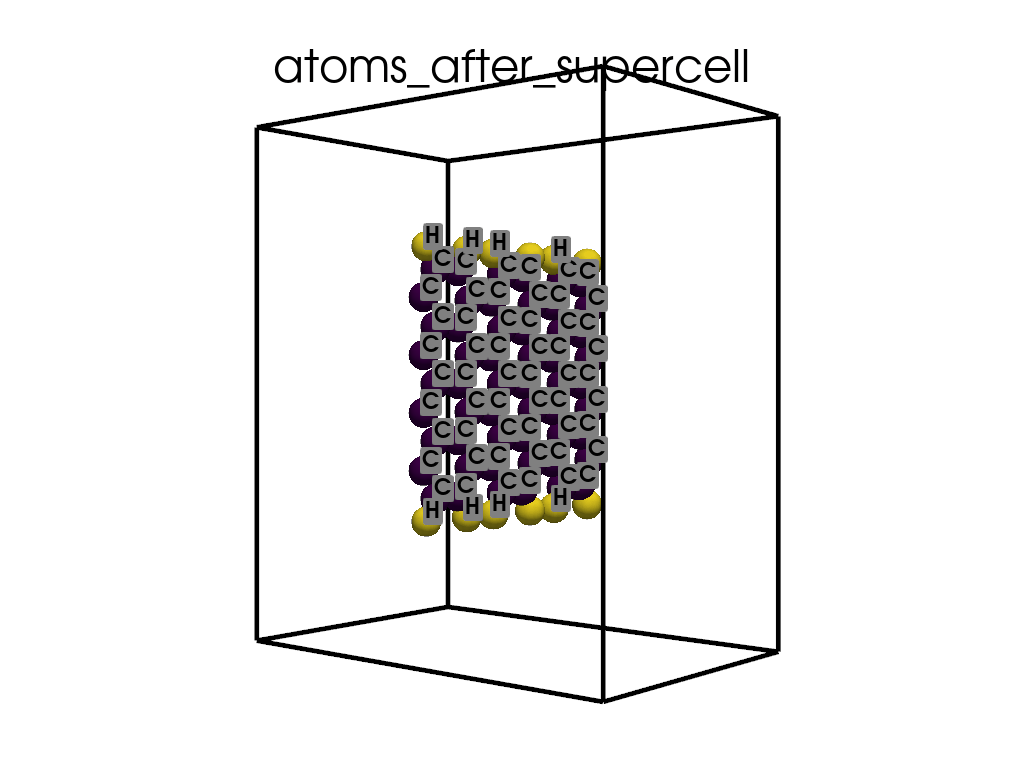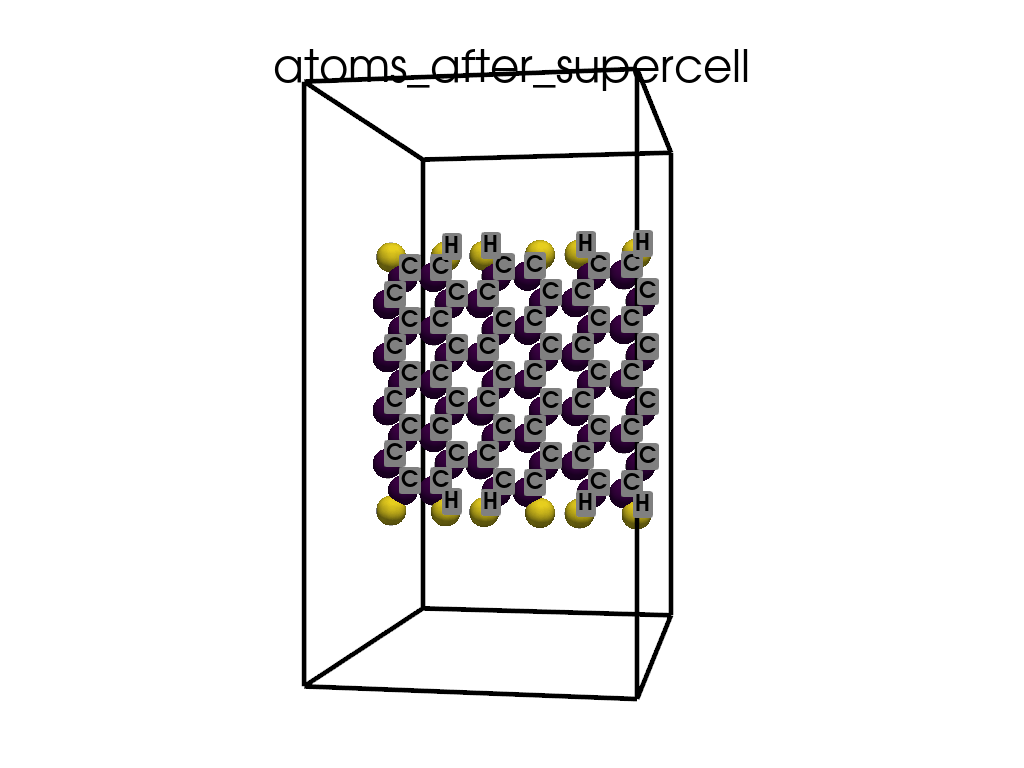
Creating a Supercell#
print("\n\nNow I will make an supercell 3x1x1")
b = p.poscarUtils.poscar_supercell(
os.path.join(data_dir, "POSCAR-9AGNR-smaller.vasp"), verbose=False
)
size = np.array([[3, 0, 0], [0, 1, 0], [0, 0, 1]])
b.supercell(size=size)
b.write(os.path.join(data_dir, "POSCAR-9AGNR-311.vasp"))
print("It was saved as POSCAR-9AGNR-311.vasp")
tmp_a = p.Poscar(os.path.join(data_dir, "POSCAR-9AGNR-smaller.vasp"))
tmp_a.parse()
# Convert positions to Cartesian coordinates for visualization
atoms_before = pv.PolyData(np.dot(tmp_a.dpos, tmp_a.lat))
atoms_before["atoms"] = tmp_a.elm
labels_before = [elm for elm, point in zip(tmp_a.elm, tmp_a.dpos)]
# Define the unit cell using lattice vectors
unit_cell_comb = list(product([0, 1], repeat=3))
unit_cell = np.array(
[
comb[0] * tmp_a.lat[0] + comb[1] * tmp_a.lat[1] + comb[2] * tmp_a.lat[2]
for comb in unit_cell_comb
]
)
unit_cell_before = pv.PolyData(unit_cell)
tmp_a = p.Poscar(os.path.join(data_dir, "POSCAR-9AGNR-311.vasp"))
tmp_a.parse()
# Convert positions to Cartesian coordinates for visualization
atoms_after = pv.PolyData(np.dot(tmp_a.dpos, tmp_a.lat))
atoms_after["atoms"] = tmp_a.elm
labels_after = [elm for elm, point in zip(tmp_a.elm, tmp_a.dpos)]
# Define the unit cell using lattice vectors
unit_cell_comb = list(product([0, 1], repeat=3))
unit_cell = np.array(
[
comb[0] * tmp_a.lat[0] + comb[1] * tmp_a.lat[1] + comb[2] * tmp_a.lat[2]
for comb in unit_cell_comb
]
)
unit_cell_after = pv.PolyData(unit_cell)
create_gif(
atoms=atoms_before,
labels=labels_before,
unit_cell=unit_cell_before,
save_file="atoms_before_supercell.gif",
)
create_gif(
atoms=atoms_after,
labels=labels_after,
unit_cell=unit_cell_after,
save_file="atoms_after_supercell.gif",
)
Now I will make an supercell 3x1x1
It was saved as POSCAR-9AGNR-311.vasp
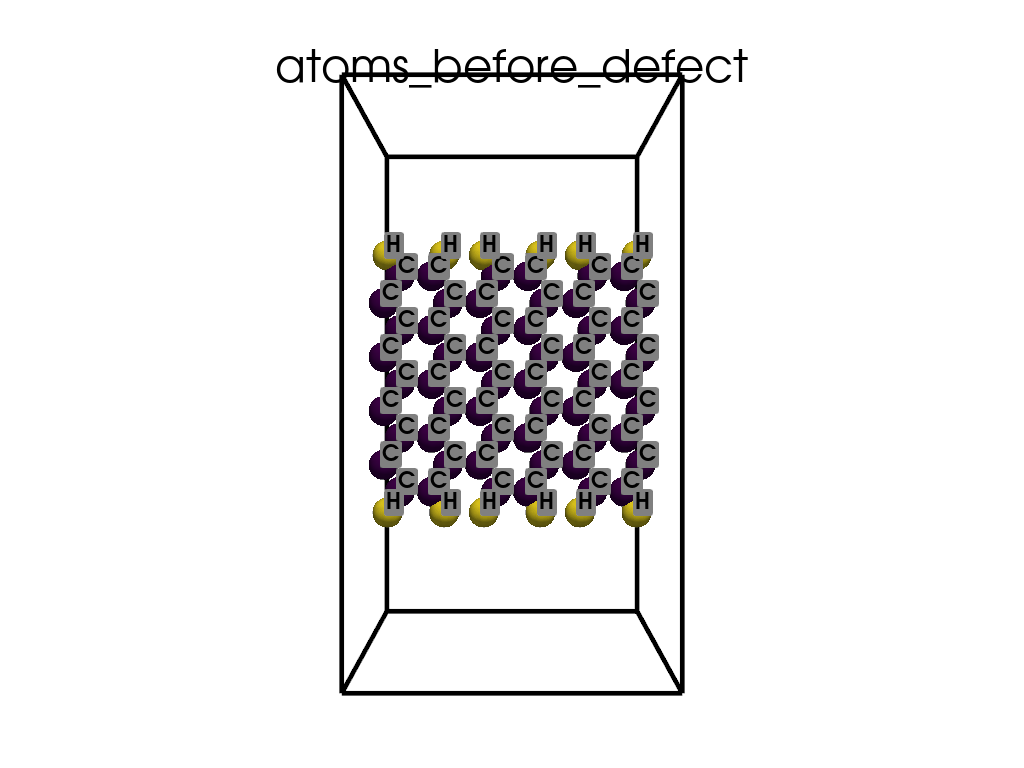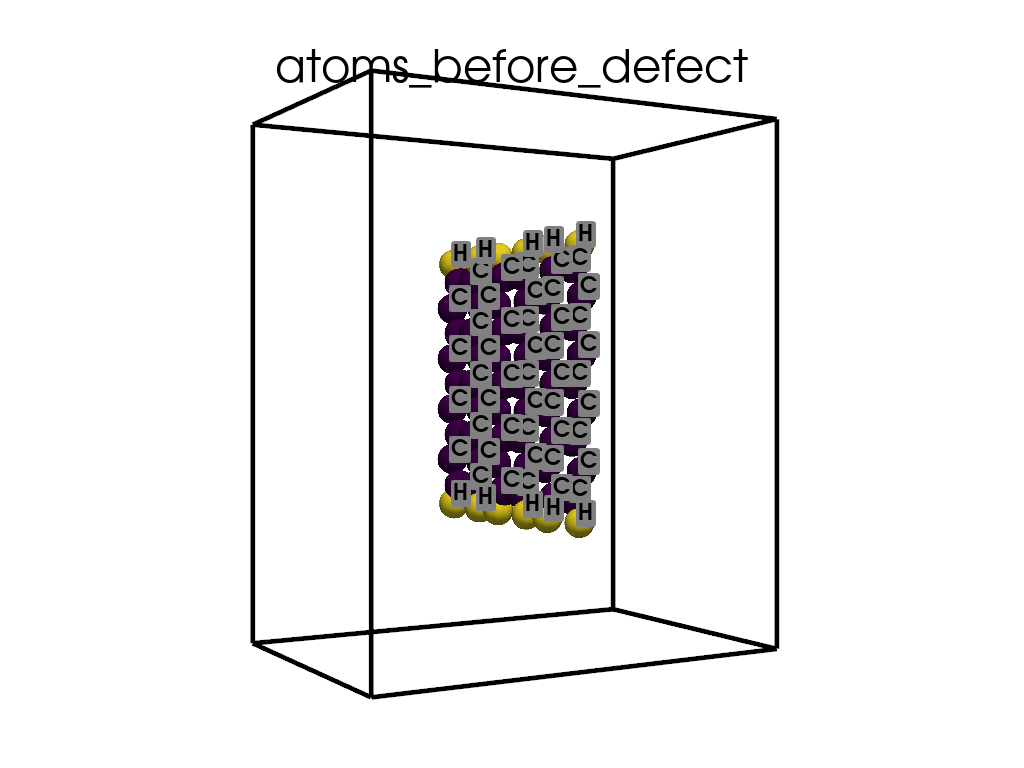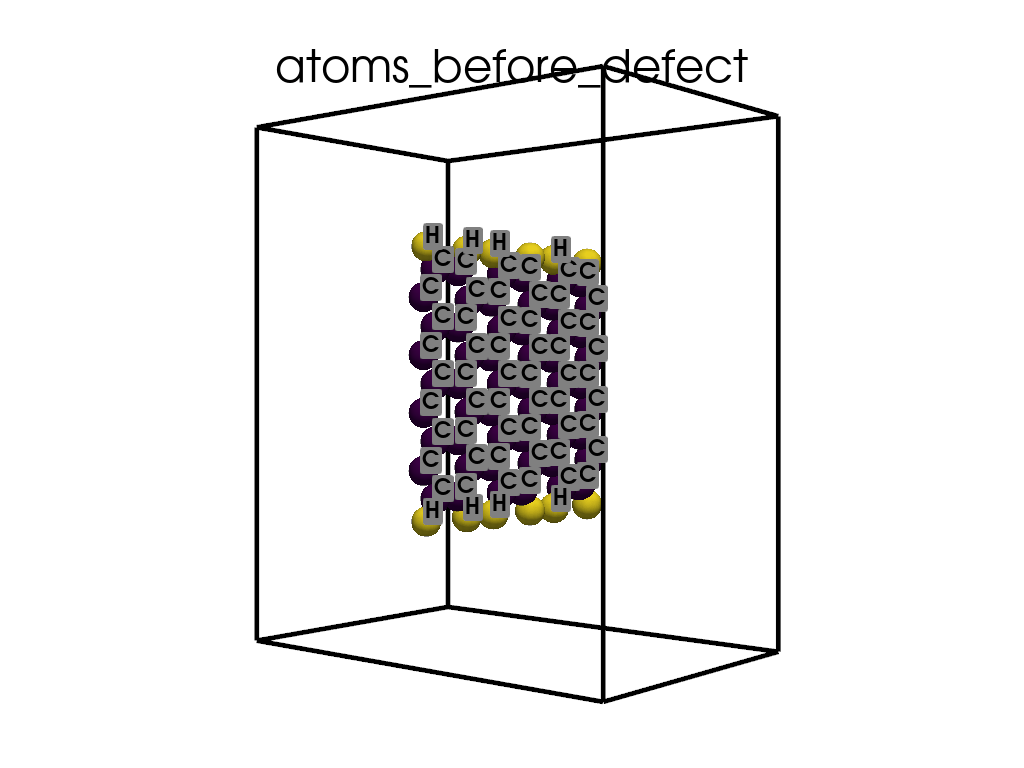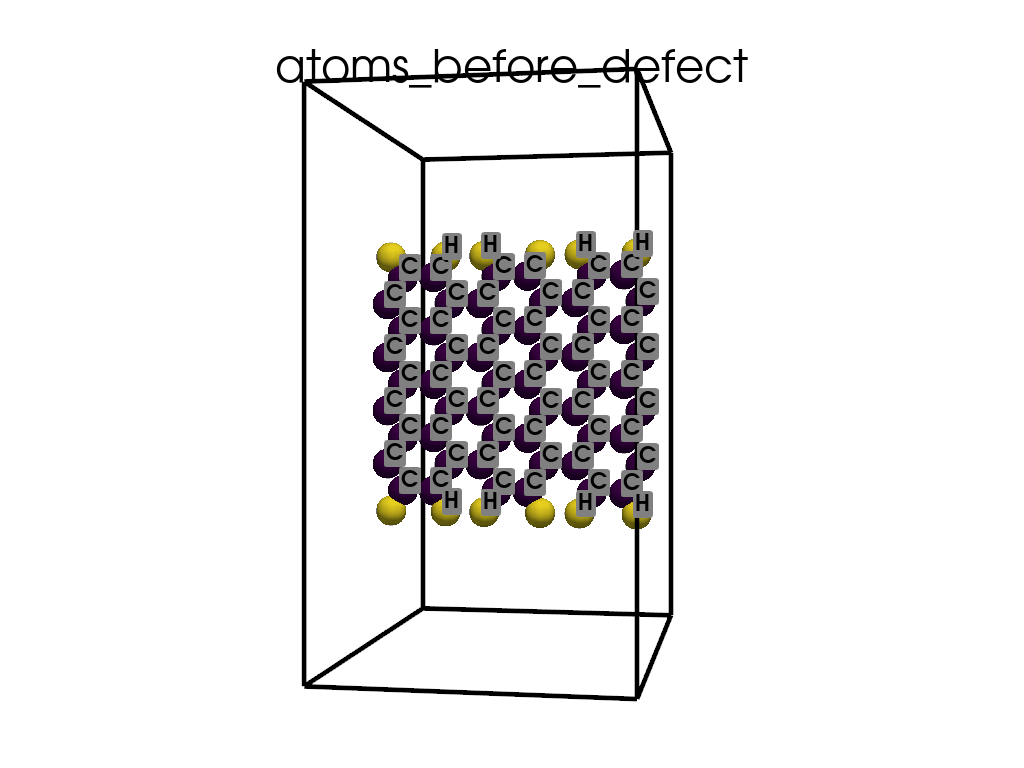
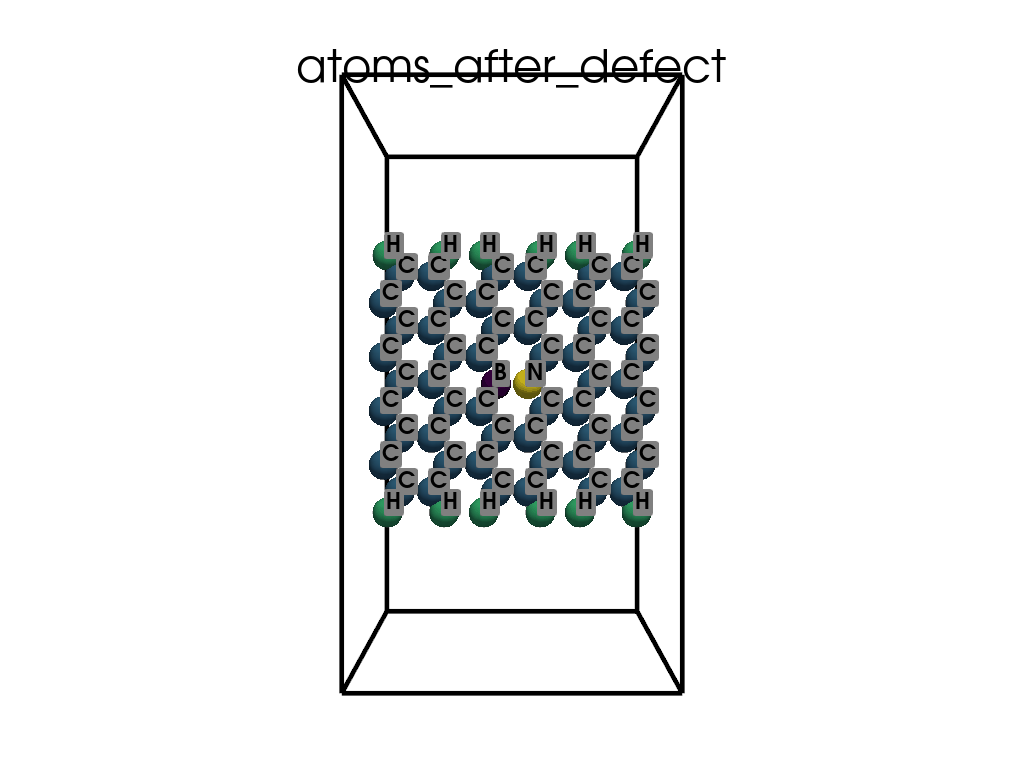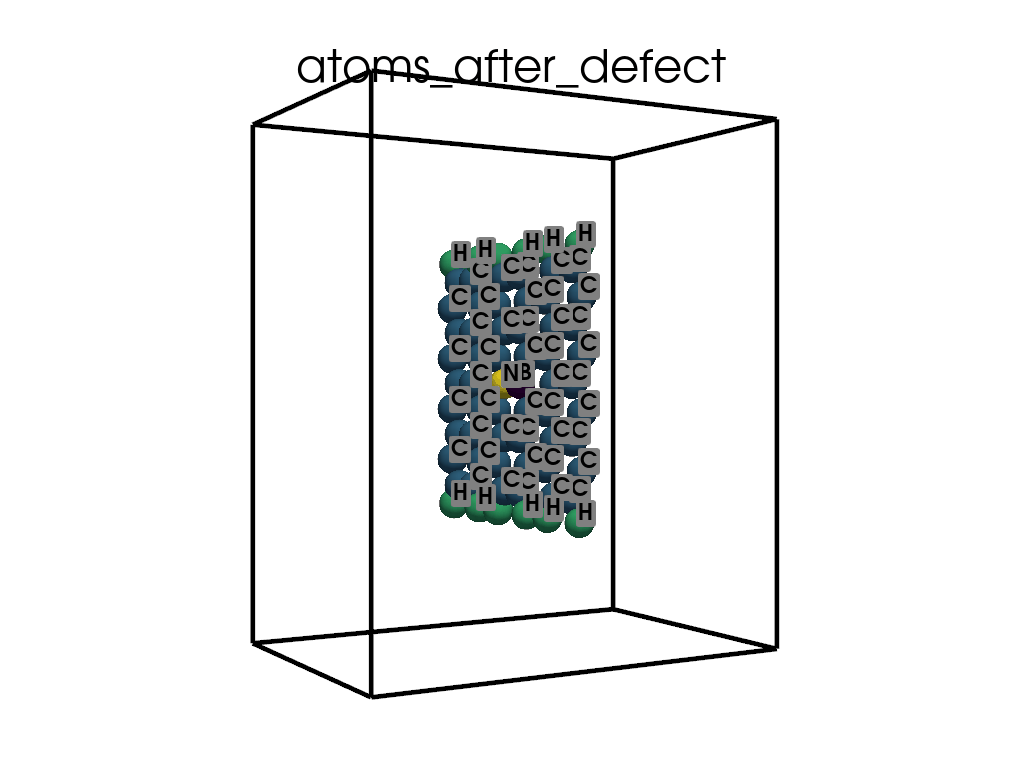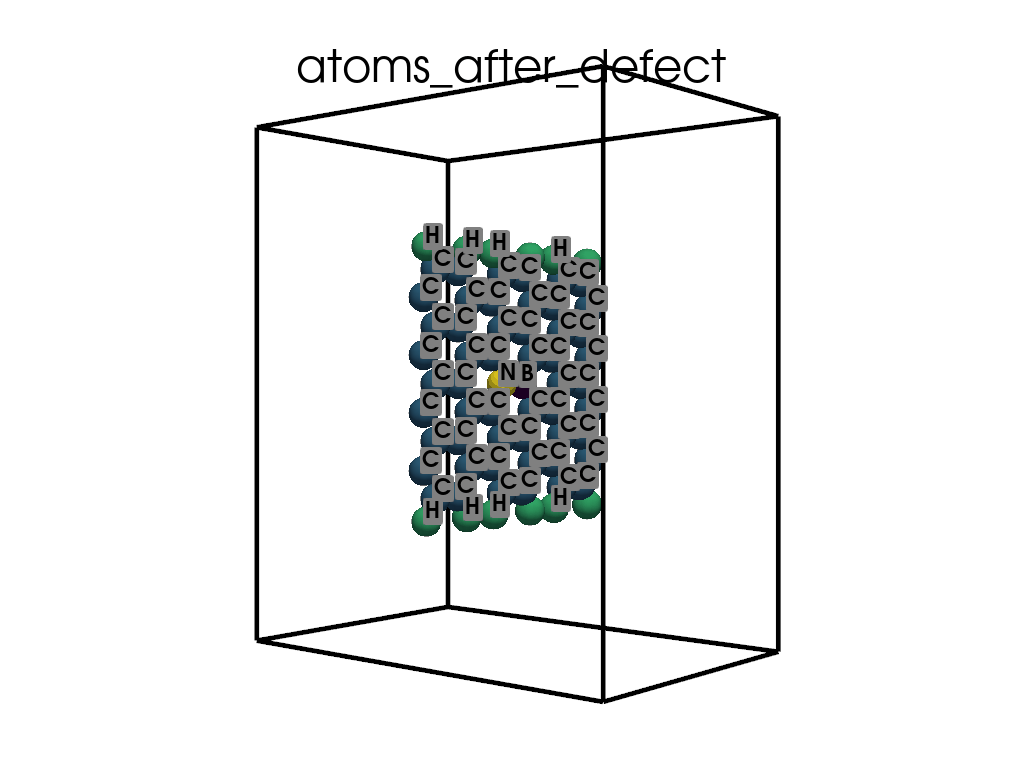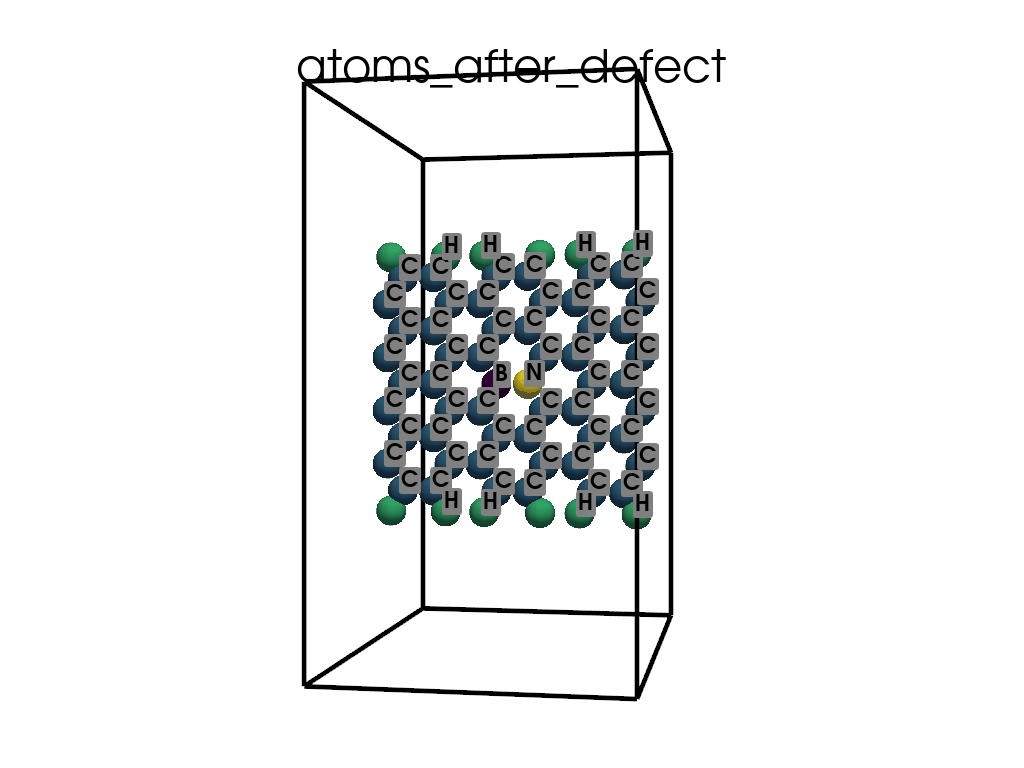
Introducing Defects#
print(
"\n\nFinally I want to create a defect by changing atoms #28, #29 to B and N, respectively"
)
c = p.poscarUtils.poscar_modify(
os.path.join(data_dir, "POSCAR-9AGNR-311.vasp"), verbose=False
)
c.change_elements(
indexes=[28, 29], newElements=["B", "N"], human=True
) # Mind, without `human`, first is 0, second is 1, ...
c.write(os.path.join(data_dir, "POSCAR-AGNR-defect.vasp"))
print("It was saves as POSCAR-AGNR-defect.vasp")
tmp_a = p.Poscar(os.path.join(data_dir, "POSCAR-9AGNR-311.vasp"))
tmp_a.parse()
# Convert positions to Cartesian coordinates for visualization
atoms_before = pv.PolyData(np.dot(tmp_a.dpos, tmp_a.lat))
atoms_before["atoms"] = tmp_a.elm
labels_before = [elm for elm, point in zip(tmp_a.elm, tmp_a.dpos)]
# Define the unit cell using lattice vectors
unit_cell_comb = list(product([0, 1], repeat=3))
unit_cell = np.array(
[
comb[0] * tmp_a.lat[0] + comb[1] * tmp_a.lat[1] + comb[2] * tmp_a.lat[2]
for comb in unit_cell_comb
]
)
unit_cell_before = pv.PolyData(unit_cell)
tmp_a = p.Poscar(os.path.join(data_dir, "POSCAR-AGNR-defect.vasp"))
tmp_a.parse()
# Convert positions to Cartesian coordinates for visualization
atoms_after = pv.PolyData(np.dot(tmp_a.dpos, tmp_a.lat))
atoms_after["atoms"] = tmp_a.elm
labels_after = [elm for elm, point in zip(tmp_a.elm, tmp_a.dpos)]
# Define the unit cell using lattice vectors
unit_cell_comb = list(product([0, 1], repeat=3))
unit_cell = np.array(
[
comb[0] * tmp_a.lat[0] + comb[1] * tmp_a.lat[1] + comb[2] * tmp_a.lat[2]
for comb in unit_cell_comb
]
)
unit_cell_after = pv.PolyData(unit_cell)
create_gif(
atoms=atoms_before,
labels=labels_before,
unit_cell=unit_cell_before,
save_file="atoms_before_defect.gif",
)
create_gif(
atoms=atoms_after,
labels=labels_after,
unit_cell=unit_cell_after,
save_file="atoms_after_defect.gif",
)
print("")
print("Loading an AGNR with a defects in the last two entries")
a = p.poscar.Poscar(os.path.join(data_dir, "POSCAR-AGNR-defect.vasp"), verbose=False)
a.parse()
print("The nearest neighbors of the defects are:")
nn = p.latticeUtils.Neighbors(a)
print(a.elm[-2], ":", a.Ntotal - 2, "-->", nn.nn_list[-2])
print(a.elm[-1], ":", a.Ntotal - 1, "-->", nn.nn_list[-1])
Finally I want to create a defect by changing atoms #28, #29 to B and N, respectively
It was saves as POSCAR-AGNR-defect.vasp
Loading an AGNR with a defects in the last two entries
The nearest neighbors of the defects are:
B : 64 --> [22, 26, 65]
N : 65 --> [25, 27, 64]
Total running time of the script: (0 minutes 22.359 seconds)